We are a bioinformatics research group at the Division of Allergy and Immunology. Our aim is to develop, improve, combine and apply bioinformatics tools to solve problems in allergy research in close collaboration with the experimental research groups.
Projects
In-Silico Exploration of Allergenic Proteins
Currently, the three-dimensional structure of about 90 allergenic proteins has been resolved experimentally. This provides a valuable source for the in-silico investigation and manipulation of such molecules. When an organism is exposed to an allergenic protein an immune response is triggered, which depends besides other factors on the three-dimensional structure of the respective allergenic protein. By the application of in-silico methods such as investigation of solvent accessibility or electrostatics, modeling of homologs and isoforms, molecular dynamics or in-silico mutagenesis in combination with experimental results we aim to gain new insights in molecular mechanisms of allergy and to support the development of new vaccines.
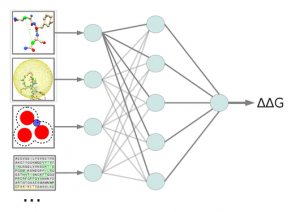
In-silico mutagenesis was successfully used to design hypoallergenic variants of the major birch pollen allergen. The approach is based on change-in-stability predictions. A current project is dedicated to improve the predictive power of the method by utilization of different machine learning algorithms in a multi-agent prediction system.
A further project is dedicated to structurally characterize allergen families by multiple structure alignments (MSTA). In order to retrieve reliable alignments a meta-alignment method was implemented, which consults different MSTA programs and combines the single method results to a consenus alignment of improved accuracy.
Prediction of Transcription Factor Binding Sites
Understanding the regulation of gene activity is a central question in biology and has high impact in medicine. Many diseases are caused by malfunctions in gene transcription control or could be treated by interfering with the molecules involved. Transcription factors (TFs) are proteins which specifically interact with the DNA and regulate the transcription of genes. The identification of potential TF binding sites on the DNA by computational methods is a long standing goal in molecular biology and supports experimentalists to hypothezise and plan experiments.